New_Genomics_Economics

1
2
3
4
5
6
7
8
9

10
11
12
13
14

15
Slide 1
 NotebookLM to PPTXTurn NotebookLM Slides into Editable PPTX
NotebookLM to PPTXTurn NotebookLM Slides into Editable PPTXExport PPTX
Log In
NotebookLM


How Deep Learning is Redefining the Cost-Benefit of Genomics
Velocity & Efficiency: The New Economics of Sequencing
NotebookLM


A single, universal, and trainable framework that achieves state-of-the-art accuracy across diverse and emerging sequencing technologies.
3.Future-Proof Adaptability


Shrink diagnostic timelines from days to hours with ultra-rapid, massively parallelized analysis pipelines, making genomics viable in acute care.
2.Clinical Velocity


Achieve higher accuracy with significantly lower sequencing coverage, directly reducing instrument and reagent costs.
1.Unprecedented Efficiency

Deep learning-based variant calling delivers a step-change in performance by fundamentally altering the trade-offs between cost, speed, and accuracy. This new paradigm enables:
From Brute Force to Intelligent Analysis
NotebookLM

Sequencing Coverage
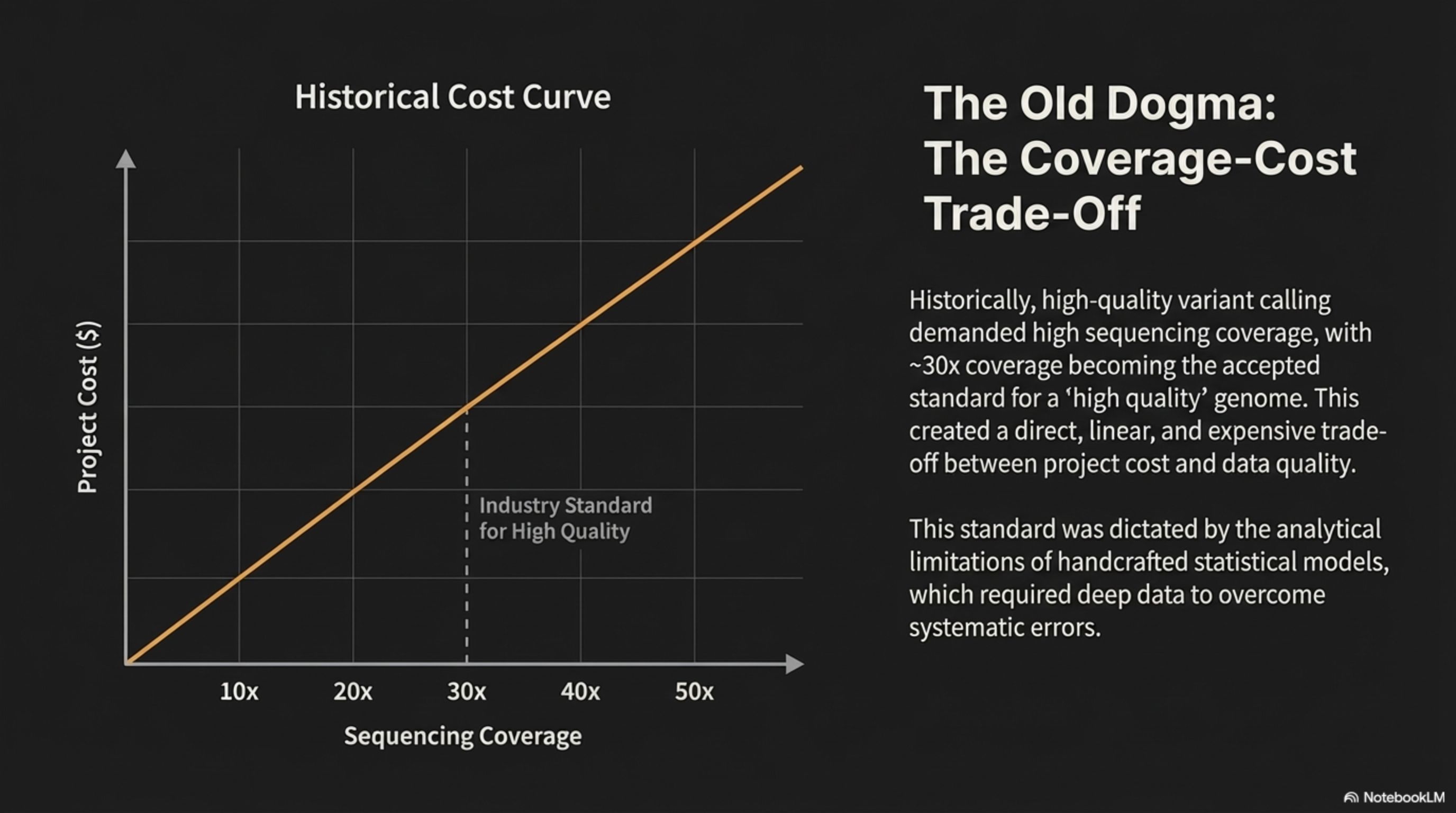
This standard was dictated by the analytical limitations of handcrafted statistical models, which required deep data to overcome systematic errors.
Historically, high-quality variant calling demanded high sequencing coverage, with ~30x coverage becoming the accepted standard for a 'high quality' genome. This created a direct, linear, and expensive trade- off between project cost and data quality.
Project Cost ($)
The Old Dogma: The Coverage-Cost Trade-Off
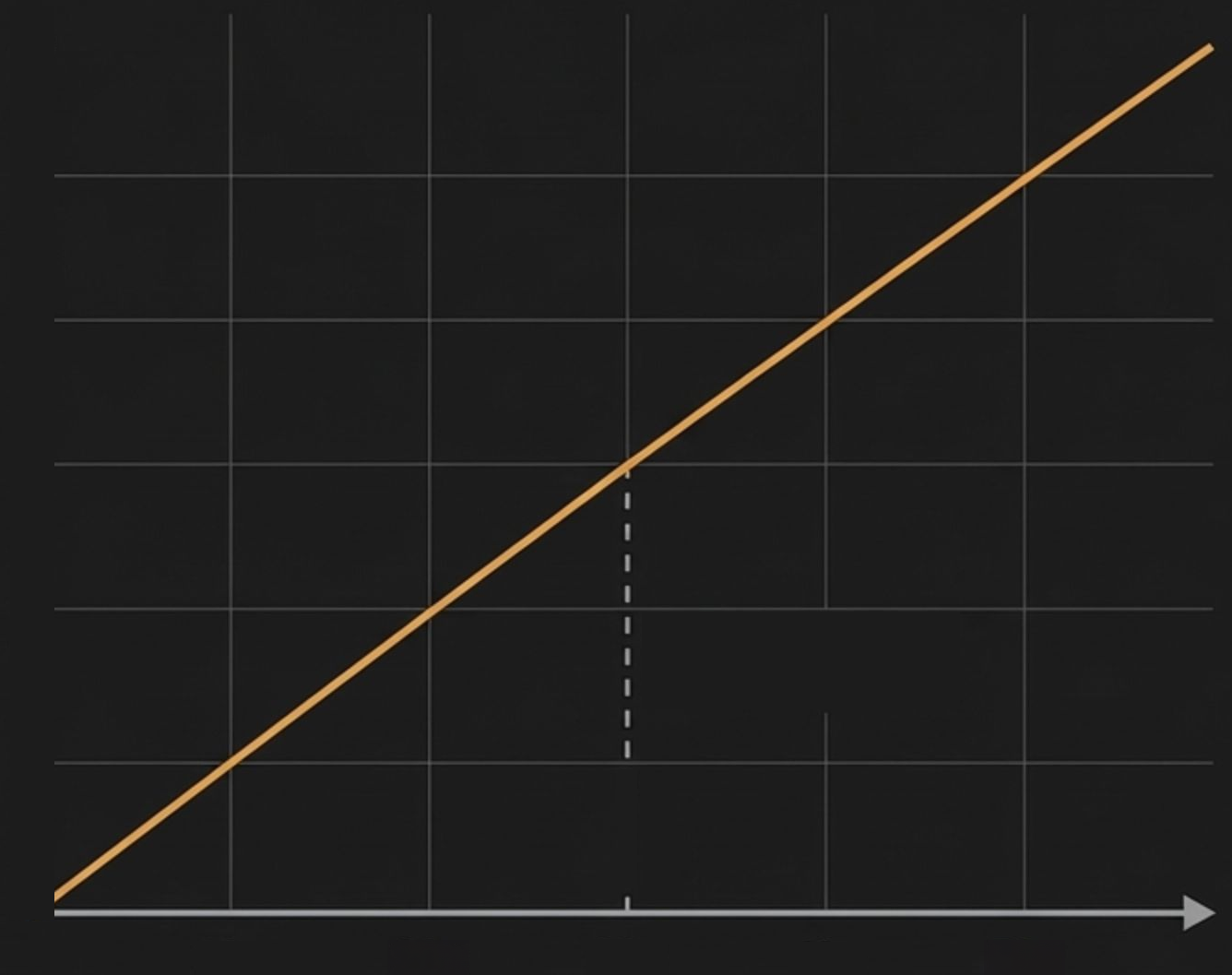
50x
40x
30x
20x
10x

Industry Standard for High Quality

Historical Cost Curve
NotebookLM

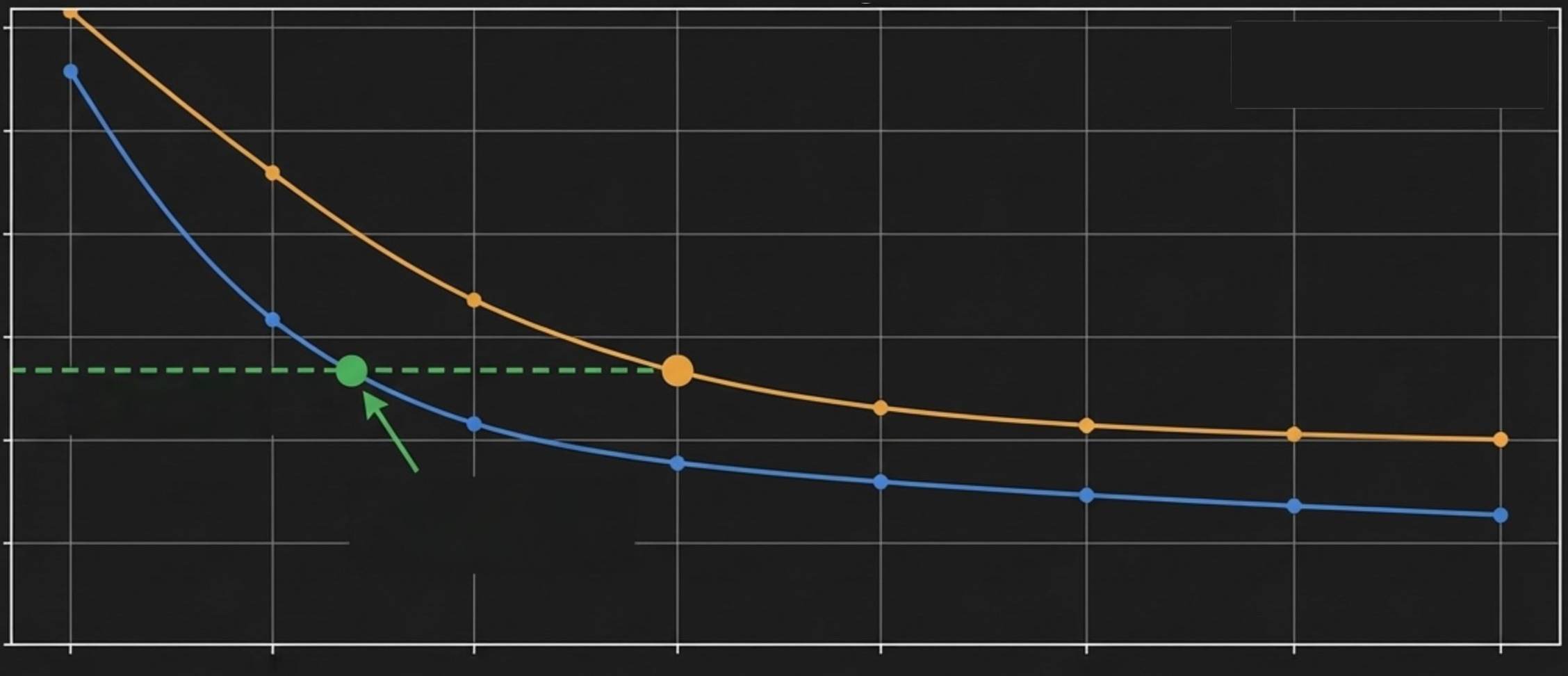
Coverage
0
25,000
50,000
75,000
100,000
TotalErrors(SNPandIndel)
125,000
150,000
50x
45x
40x
35x
30x
25x
20x
1Sx
**Equivalent Accuracy Achievedat~22x Coverage.**
at 30x Coverage
Errors at GATK4.1-HC
GATK4.1.0.0
DeepVariant v0.8.0
Total Errors vs. Coverage on HG002
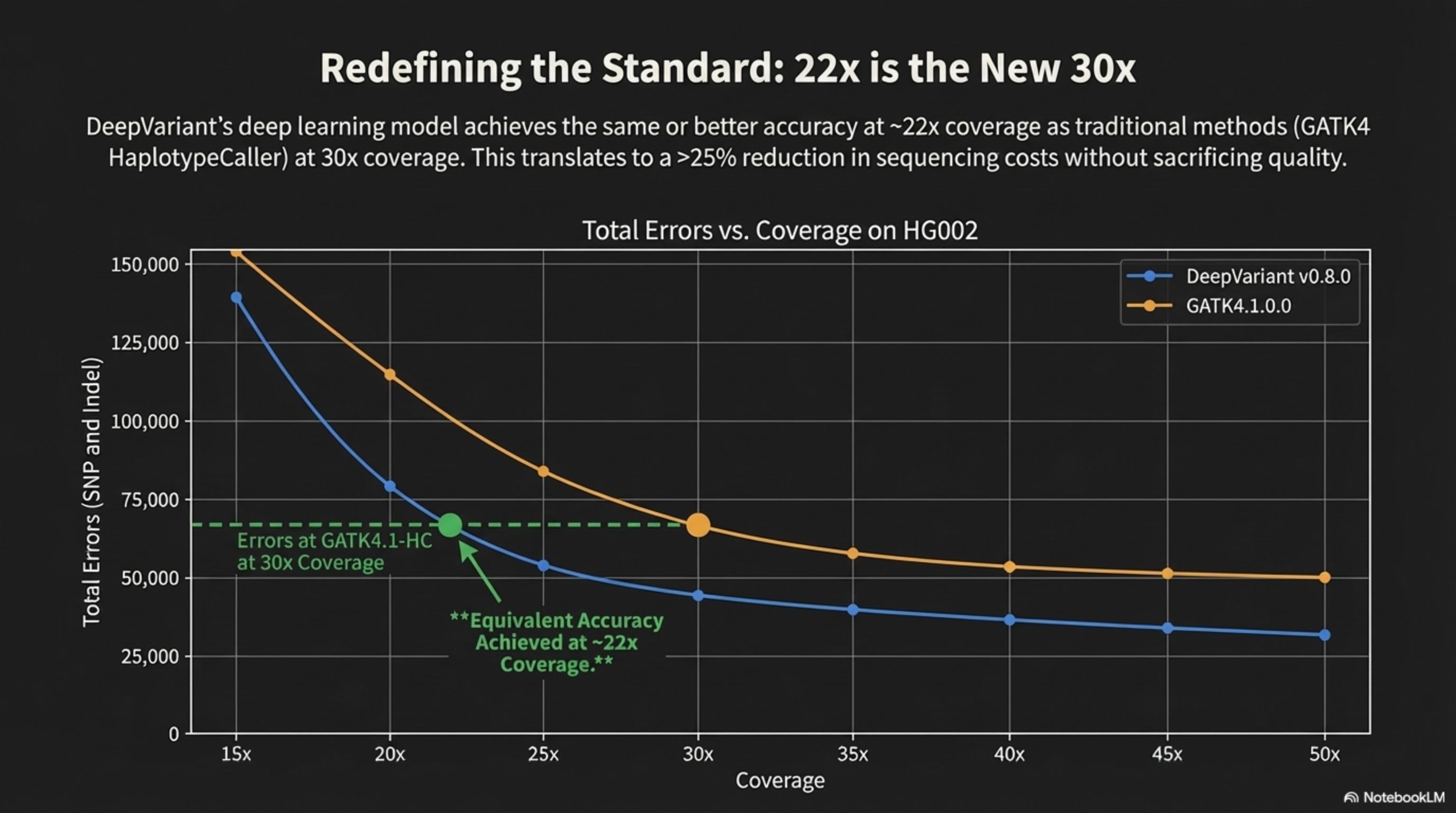
DeepVariant's deep learning model achieves the same or better accuracy at ~22x coverage as traditional methods (GATK4 HaplotypeCaller) at 30x coverage. This translates to a >25% reduction in sequencing costs without sacrificing quality.
Redefining the Standard: 22x is the New 30x
NotebookLM

Coverage of each sample in trio
35x
30x
25x
20x
15x
0.986
0.988
0.980
0.992
0.994
OverallFlScore
0.996
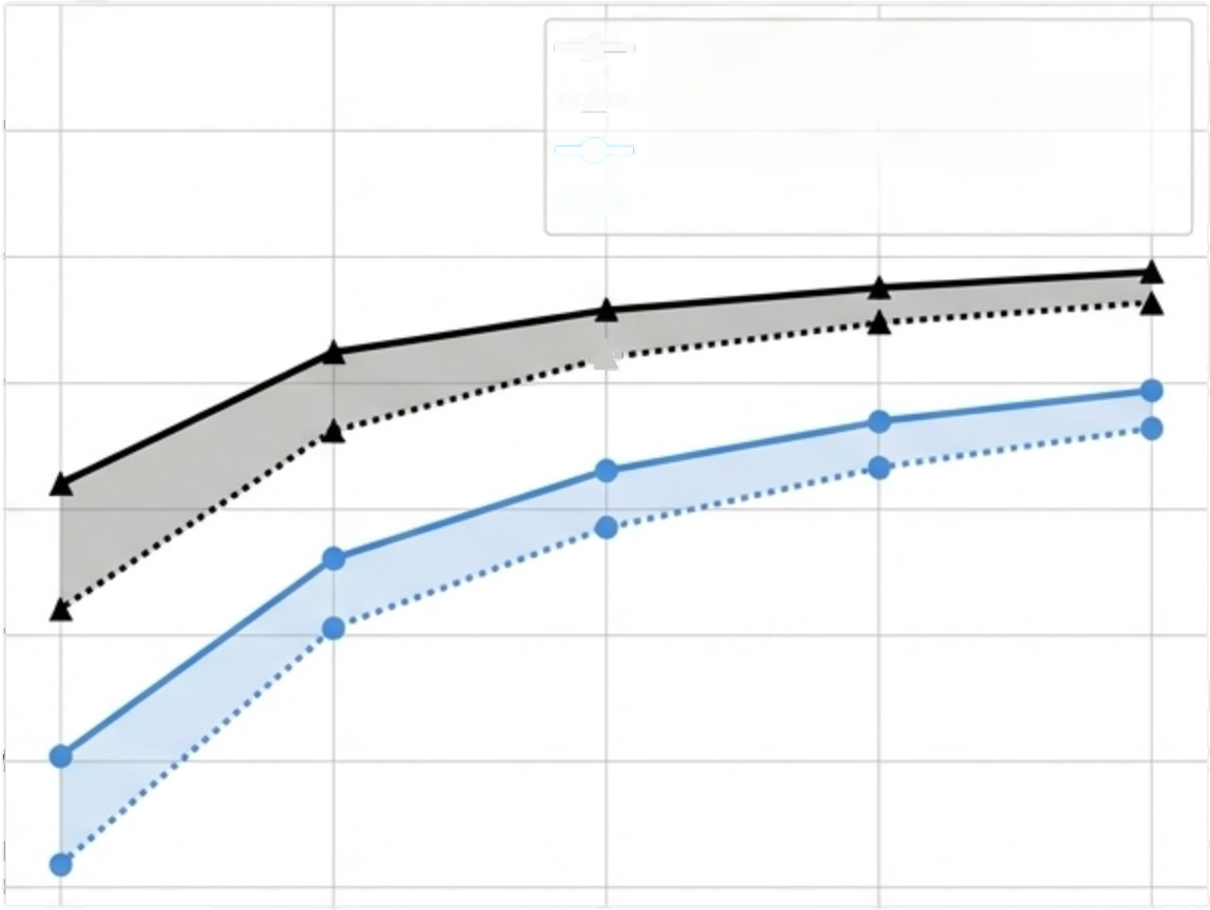
DeepVariant Illumina (non-trio)
DeepTrio Illumina (trio)

DeepVariant PacBio (non-trio)
DeepTrio PacBio (trio)


0.998
F1 Score vs. Coverage for HG002( (Child)
1.000
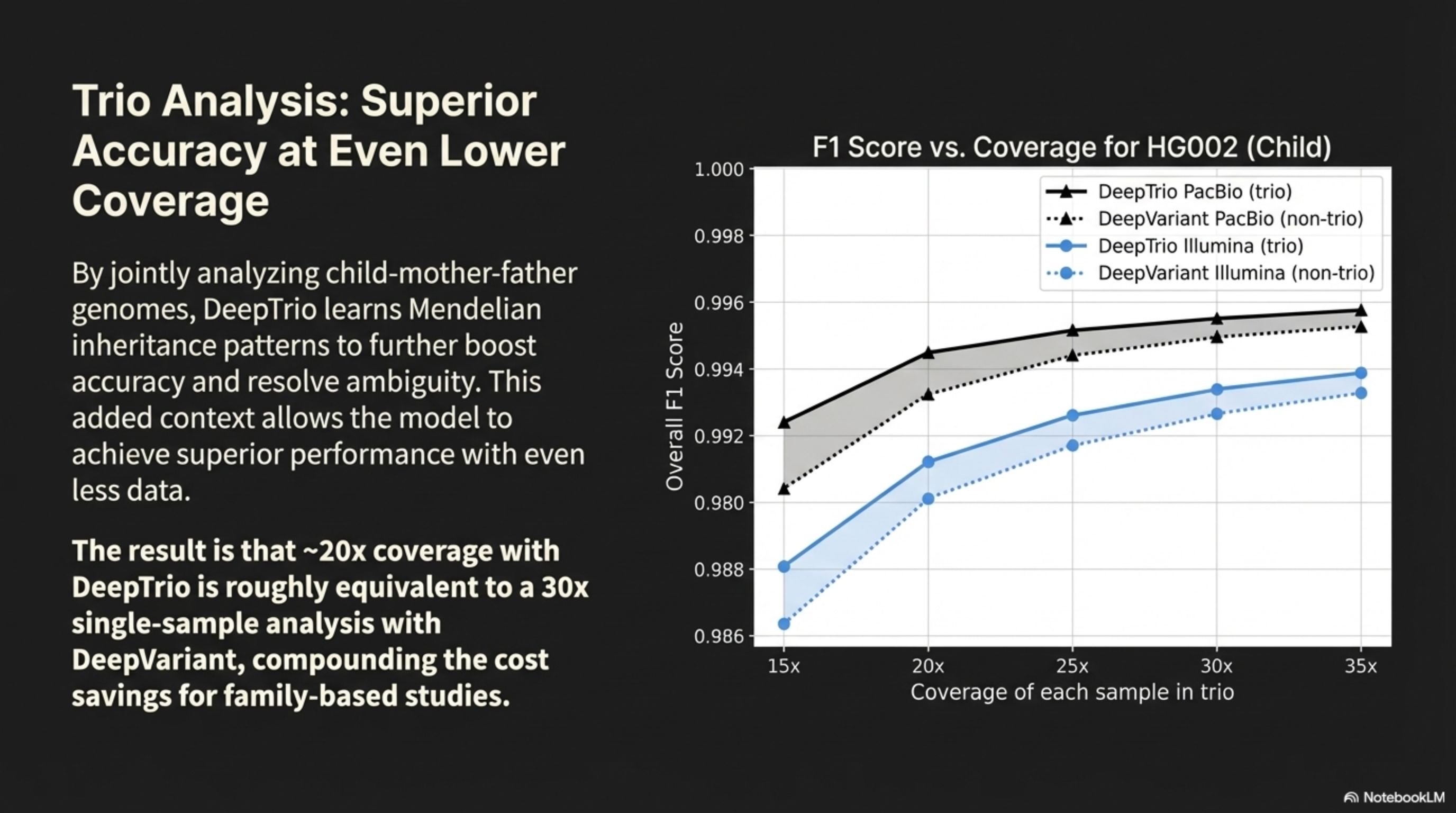
The result is that ~20x coverage with DeepTrio is roughly equivalent to a 30x single-sample analysis with DeepVariant, compounding the cost savings for family-based studies.
By jointly analyzing child-mother-father genomes, DeepTrio learns Mendelian inheritance patterns to further boost accuracy and resolve ambiguity. This added context allows the model to achieve superior performance with even less data.
Trio Analysis: Superior Accuracy at Even Lower Coverage
NotebookLM








read supports base differs variantfrom ref
strand
quality
mapping
base quality
read base
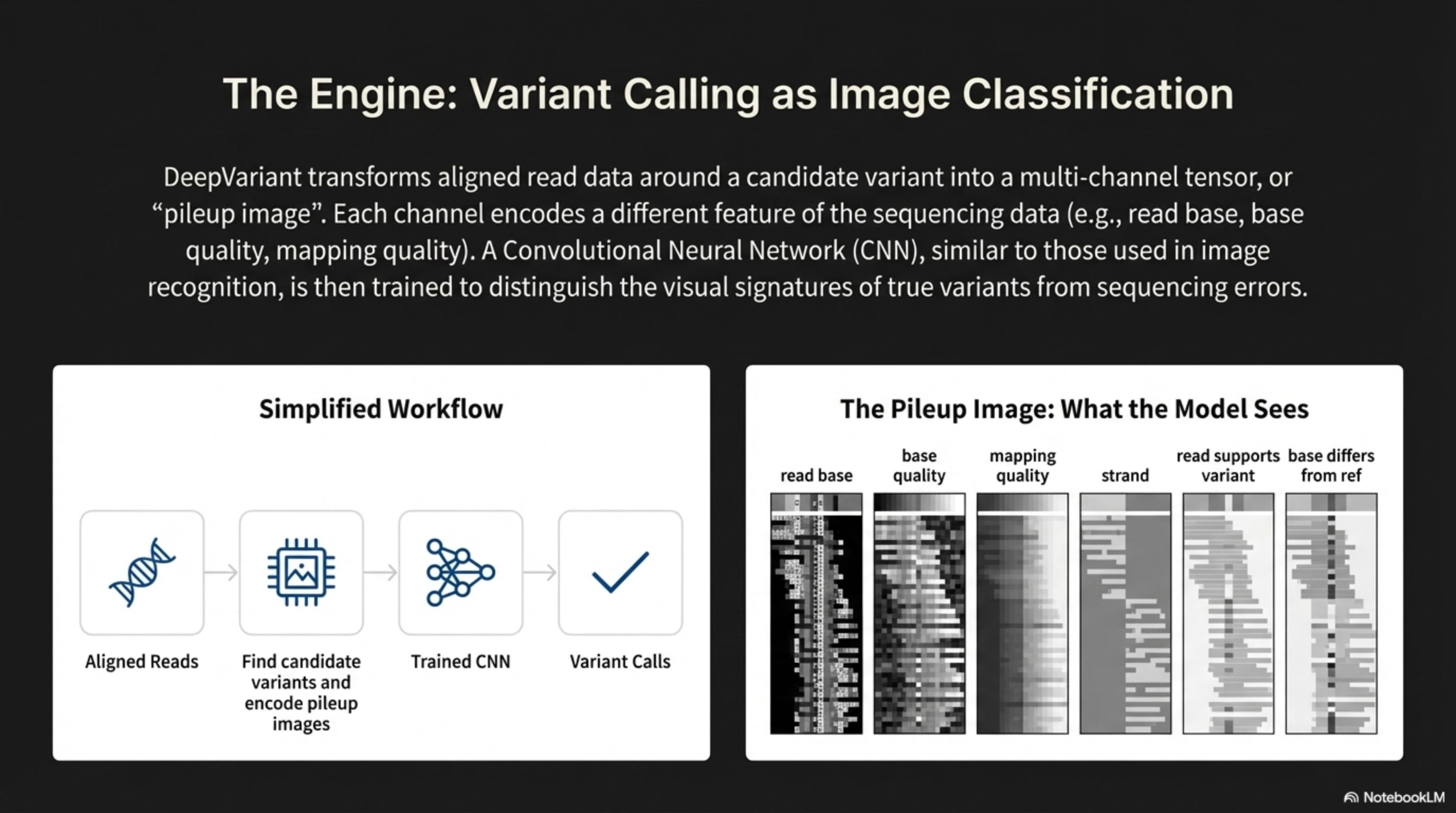
The Pileup Image: What the Model Sees
Variant Calls

Trained CNN



Find candidate variants and encode pileup images



Aligned Reads


Simplified Workflow
DeepVariant transforms aligned read data around a candidate variant into a multi-channel tensor, or "pileup image". Each channel encodes a different feature of the sequencing data (e.g,, read base, base quality, mapping quality). A Convolutional Neural Network (CNN), similar to those used in image recognition, is then trained to distinguish the visual signatures of true variants from sequencing errors.
The Engine: Variant Calling as Image Classification
NotebookLM


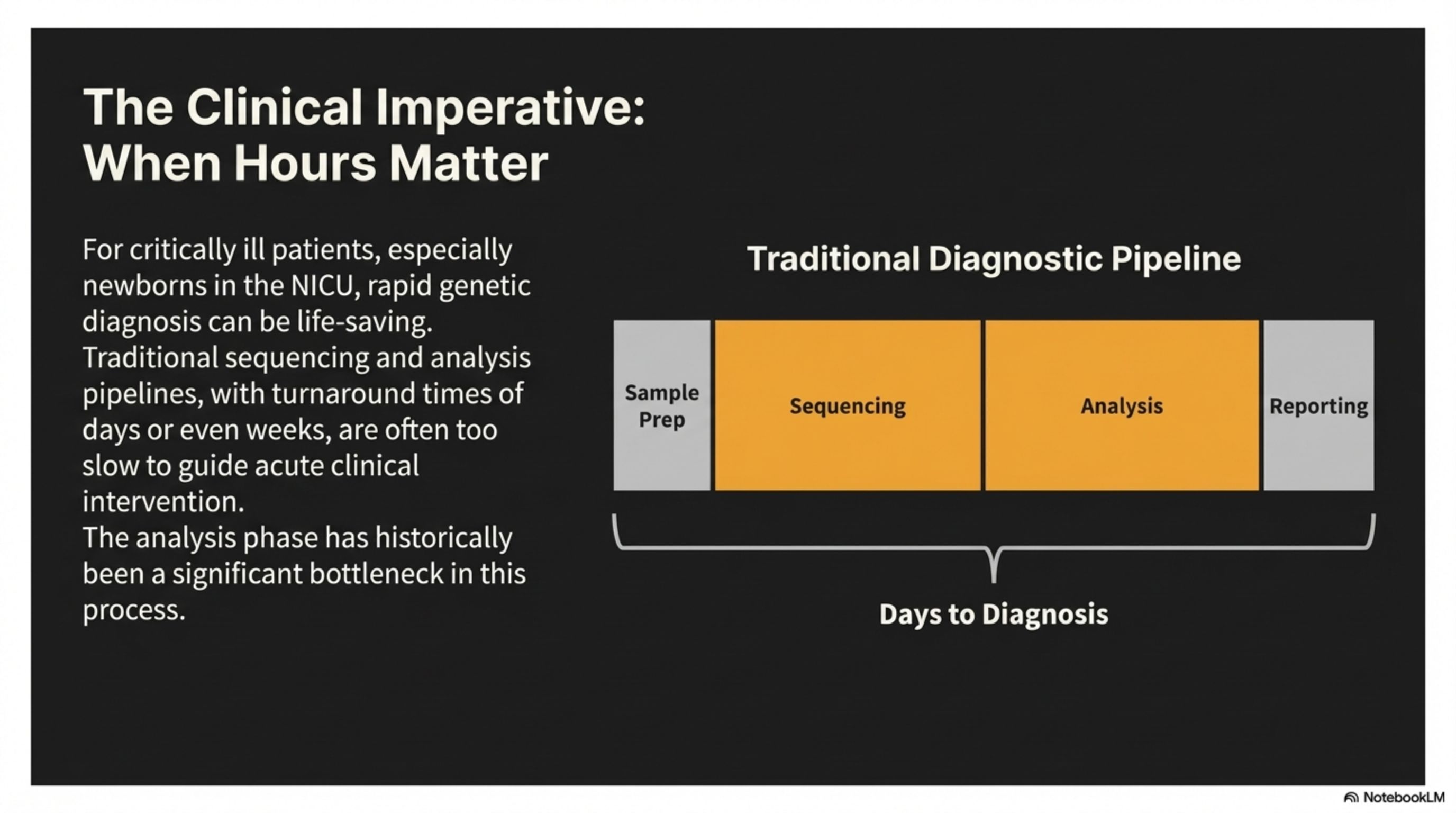
Days to Diagnosis
Reporting
Analysis
Sequencing

Sample Prep
Traditional Diagnostic Pipeline
For critically ill patients, especially newborns in the NicU, rapid genetic diagnosis can be life-saving. Traditional sequencing and analysis pipelines, with turnaround times of days or even weeks, are often too slow to guide acute clinical intervention. The analysis phase has historically been a significant bottleneck in this process.
The Clinical Imperative: When Hours Matter
NotebookLM



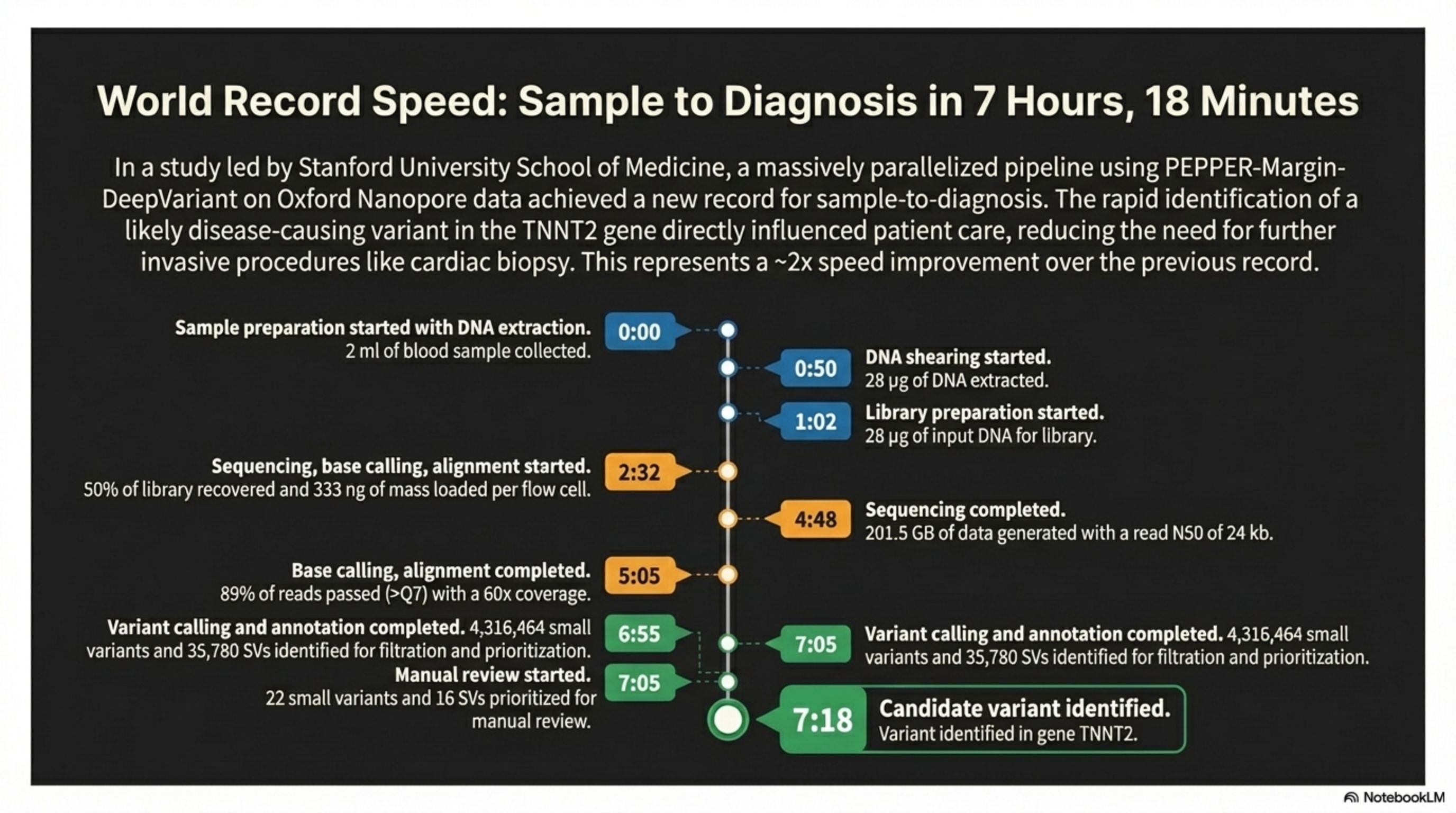
Variant identified in gene TNNT2.
Candidate variant identified.
7:18
variants and 35,780 SVs identified for filtration and prioritization.

7:05
Variant calling and annotation completed.

7:05

6:55
Variant calling and annotation completed.4,316,464 small variants and 3S,780 SVs identified for fiitration and prioritization. Manual review started. 22 small variants and 16 SVs prioritized for manual review.

5:05
89% of reads passed (>Q7) with a 60x coverage.
Base calling, alignment completed.
201.5 GB of data generated with a read NS0 of 24 kb.
Sequencing completed.
4:48
2:32
50% of library recovered and 333 ng of mass loaded per flow cell.
Sequencing, base calling, alignment started.
28ug of input DNA for library.
Library preparation started.

1:02
28 pg of DNA extracted.
DNA shearing started.

0:50



0:00
2 ml of blood sample collected.
Sample preparation started with DNA extraction.
In a study led by Stanford University School of Medicine, a massively parallelized pipeline using PEPPER-Margin- DeepVariant on Oxford Nanopore data achieved a new record for sample-to-diagnosis. The rapid identification of a likely disease-causing variant in the TNNT2 gene directly influenced patient care, reducing the need for further invasive procedures like cardiac biopsy. This represents a ~2x speed improvement over the previous record.
World Record Speed: Sample to Diagnosis in 7 Hours, 18 Minutes
NotebookLM

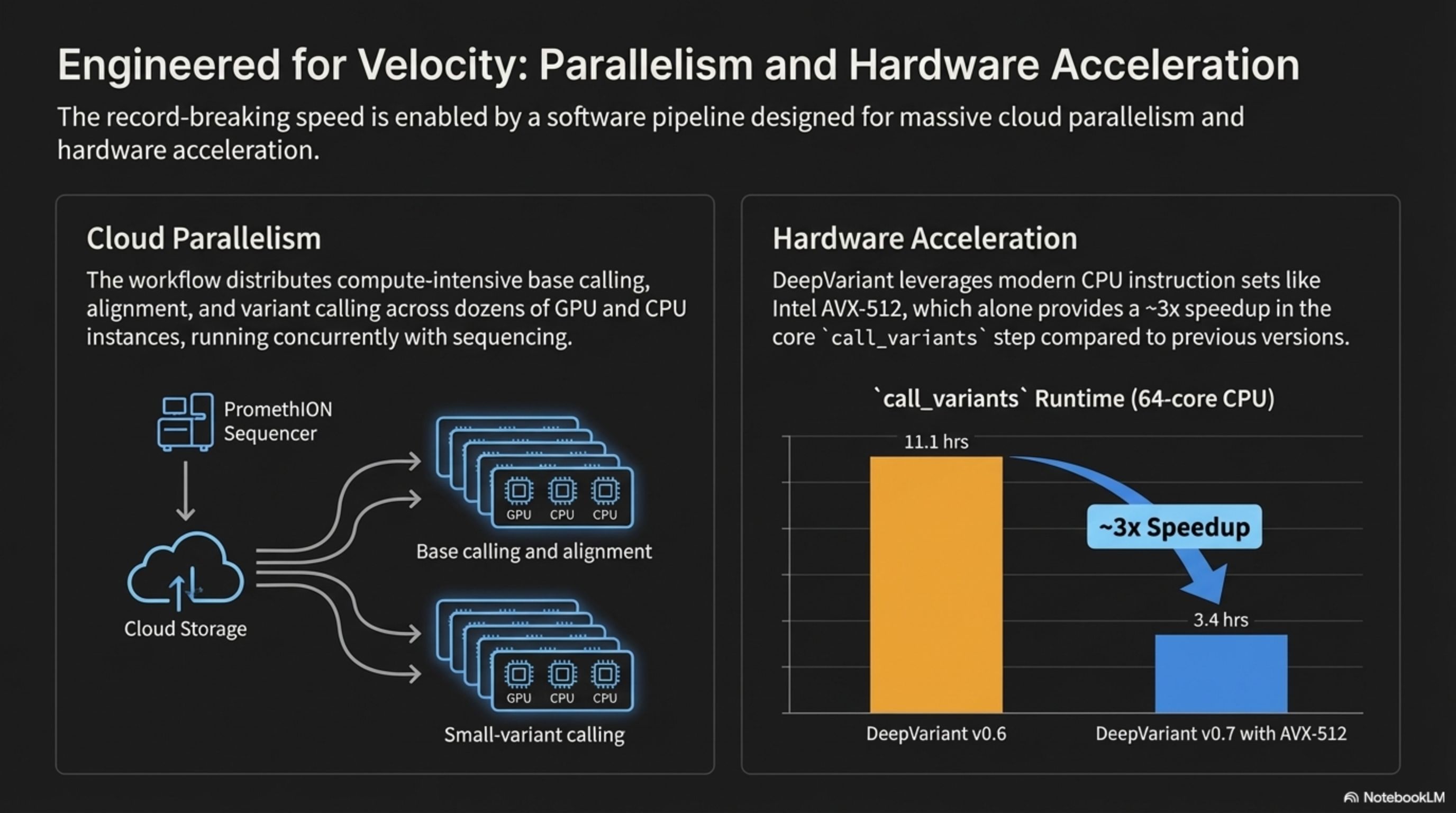
DeepVariant v0.7 with AVX-512
DeepVariant v0.6


3.4hrs
~3x Speedup
11.1hrs
`call_variants` Runtime (64-core CPU)
DeepVariant leverages modern CPU instruction sets like Intel Avx-512, which alone provides a ~3x speedup in the core `call_variants step compared to previous versions.
Hardware Acceleration

Small-variant calling

CPU
CPU
GPU



Cloud Storage
CPU
CPU
GPU



Base calling and alignment

Sequencer
PromethlON

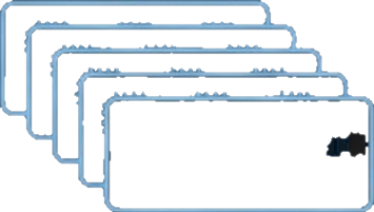
The workflow distributes compute-intensive base calling, alignment, and variant calling across dozens of GPU and CPU instances, running concurrently with sequencing
Cloud Parallelism
The record-breaking speed is enabled by a software pipeline designed for massive cloud parallelism and hardware acceleration.
Engineered for Velocity: Parallelism and Hardware Acceleration
NotebookLM




NANOPORE
OXFORD




Bio
Pac



llumina°
i
Unlike traditional statistical models that are finely tuned for a specific technology's error profile, DeepVariant's learning-based framework is technology-agnostic. By re-training on new, representative data, it achieves state-of-the-art accuracy across all major sequencing platforms.
A Universal Caller for a Multi-Platform World

NotebookLM
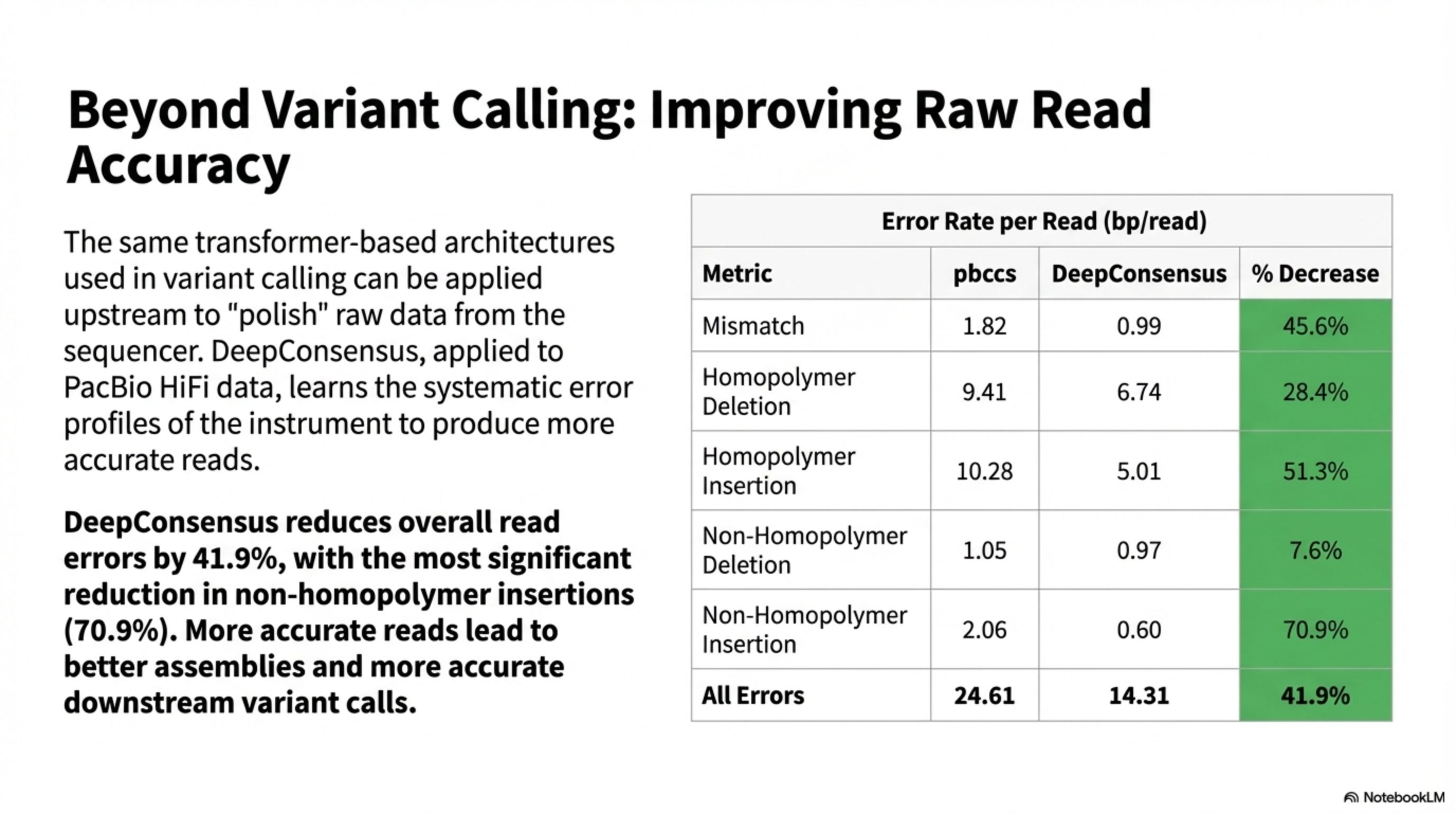
DeepConsensus reduces overall read errors by 41.9%, with the most significant reduction in non-homopolymer insertions (70.9%). More accurate reads lead to better assemblies and more accurate downstream variant calls.
The same transformer-based architectures used in variant calling can be applied upstream to "polish" raw data from the sequencer. DeepConsensus, applied to PacBio HiFi data, learns the systematic error profiles of the instrument to produce more accurate reads.
41.9%
14.31
24.61
All Errors
70.9%
0.60
2.06
Non-Homopolymer Insertion
7.6%
0.97
1.05
Non-Homopolymer Deletion
51.3%
5.01
10.28
Homopolymer Insertion
28.4%
6.74
9.41
Homopolymer Deletion
45.6%
0.99
1.82
Mismatch
% Decrease
DeepConsensus
pbccs
Metric
Error Rate per Read (bp/read)
Beyond Variant Calling: Improving Raw Read Accuracy
NotebookLM

Source Sans Pro SemiBold
Baseline + HP Channel
Baseline


Reduction
INDEL Error
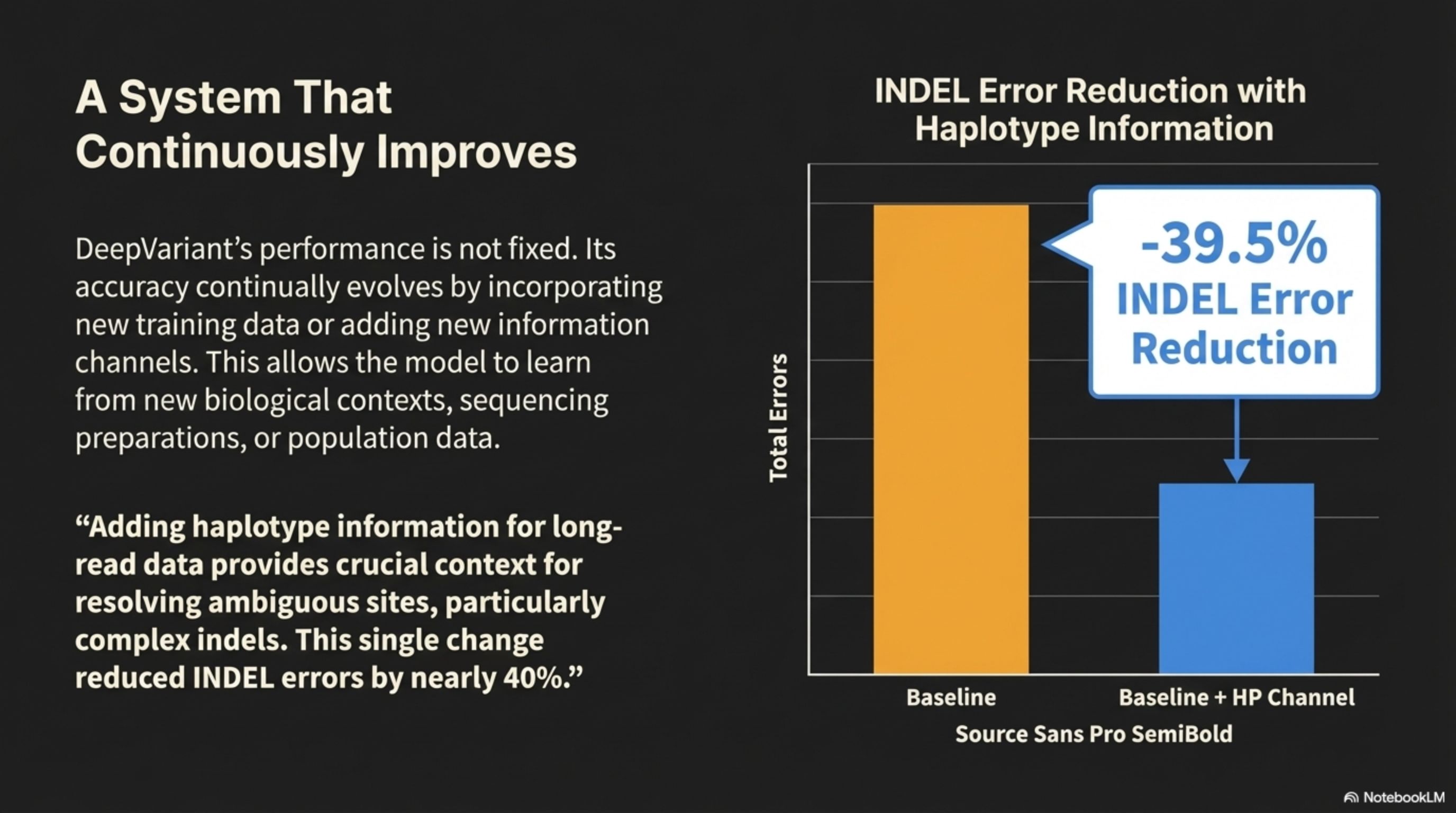
-39.5%
"Adding haplotype information for long- read data provides crucial context for resolving ambiguous sites, particularly complex indels. This single change reduced INDEL errors by nearly 40%."
DeepVariant's performance is not fixed. Its accuracy continually evolves by incorporating new training data or adding new information channels. This allows the model to learn from new biological contexts, sequencing preparations, or population data.
INDEL Error Reduction with Haplotype Information
A System That Continuously Improves

NotebookLM
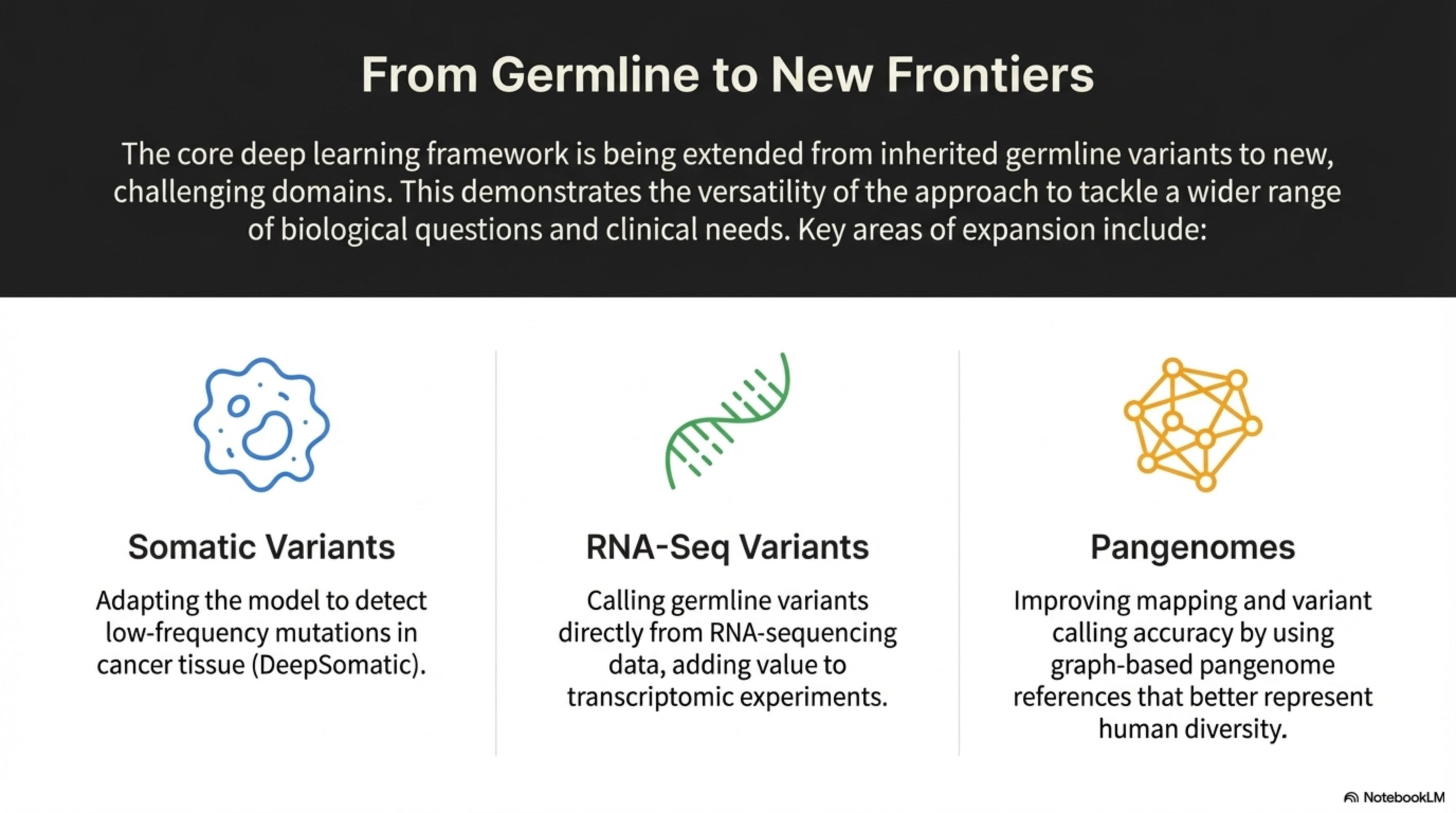
Improving mapping and variant calling accuracy by using graph-based pangenome references that better represent human diversity.
Pangenomes

Calling germline variants directly from RNA-sequencing data, adding value to transcriptomic experiments.
RNA-Seq Variants


Adapting the model to detect low-frequency mutations in cancer tissue (DeepSomatic).
Somatic Variants


The core deep learning framework is being extended from inherited germline variants to new, challenging domains. This demonstrates the versatility of the approach to tackle a wider range of biological questions and clinical needs. Key areas of expansion include:
From Germline to New Frontiers


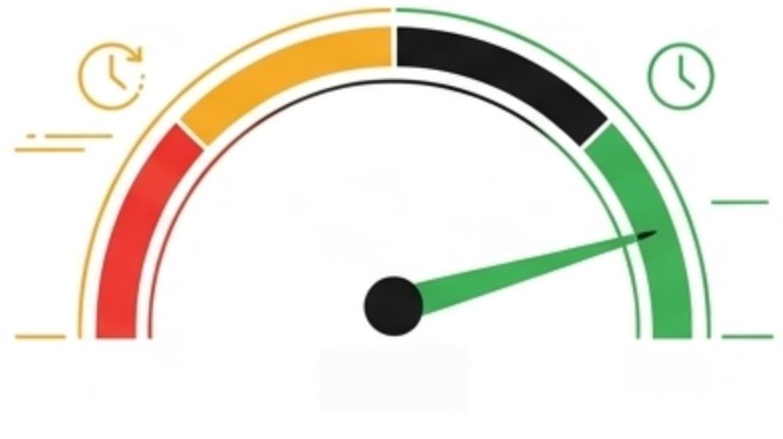
High
HIGH
Low
Source Sans Pro SemiBold
Accuracy



Low
<8hrs
High
Source Sans Pro SemiBold
Time to Diagnosis
High
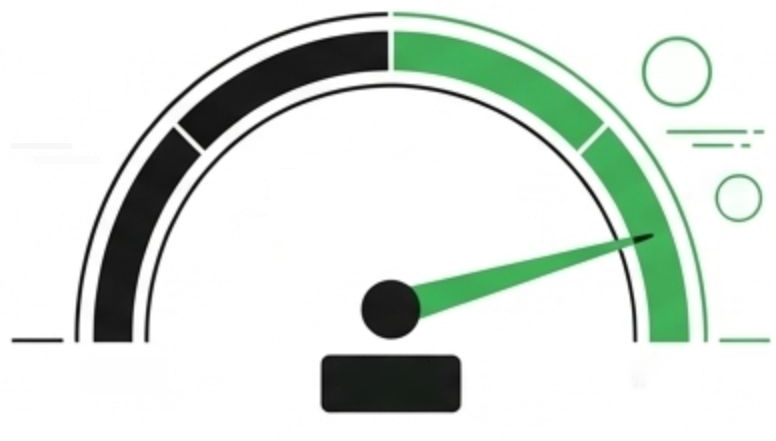
HIGH
Low
Source Sans Pro SemiBold
$
Cost Savings
$


$
$
NotebookLM

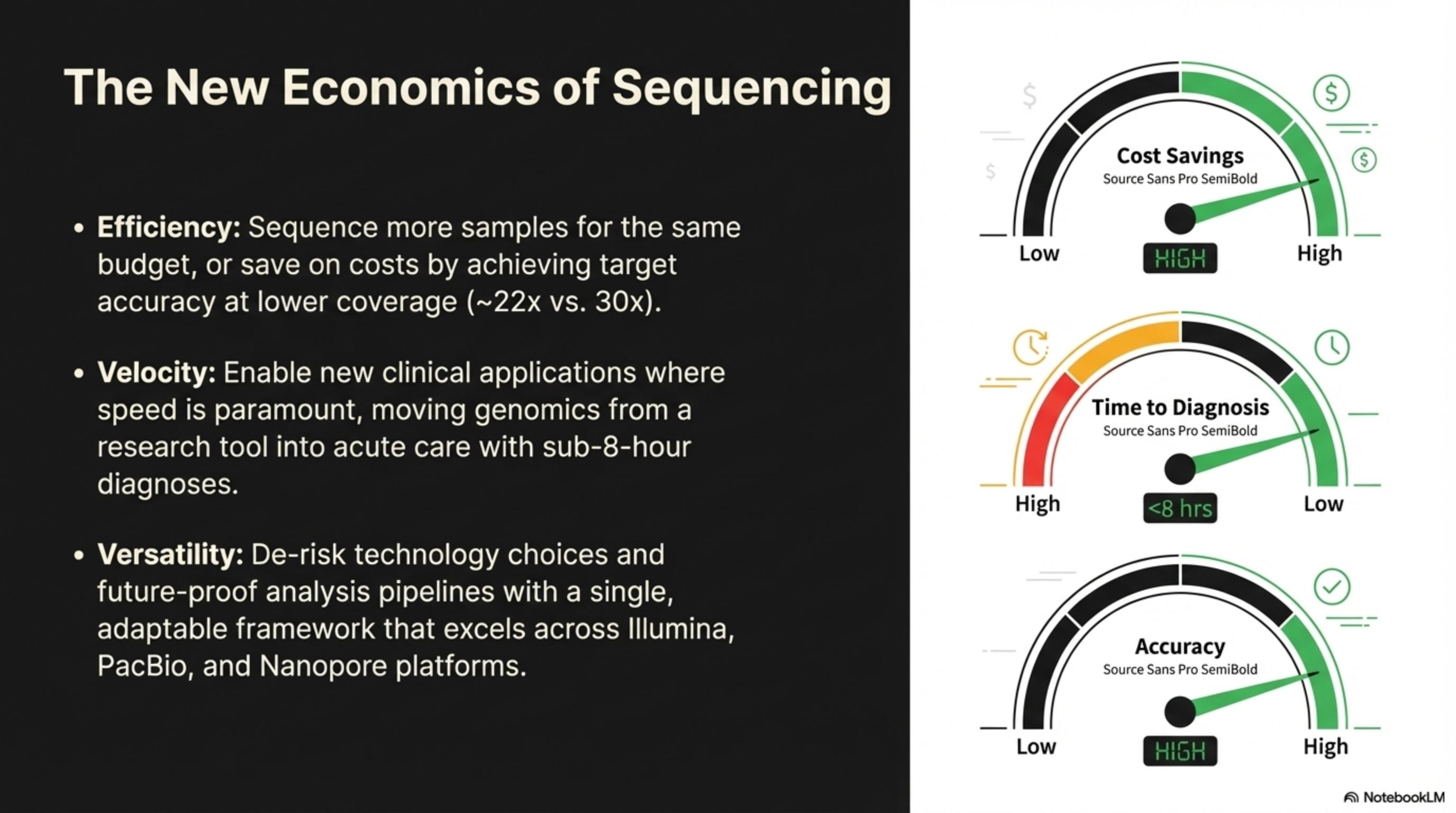
Versatility: De-risk technology choices and future-proof analysis pipelines with a single, adaptable framework that excels across Illumina, PacBio, and Nanopore platforms.

speed is paramount, moving genomics from a research tool into acute care with sub-8-hour diagnoses.
Enable new clinical applications where
Velocity:

Sequence more samples for the same
budget, or save on costs by achieving target accuracy at lower coverage (~22x vs. 30x).
Efficiency:

The New Economics of Sequencing
NotebookLM

Kolesnikov, A. et al. DeepTrio: Variant Calling in Families Using Deep Learning. bioRxiv (2021).

sequencing. Nat Biotechnol (2022).
Goenka, S.D. et al. Accelerated identification of disease-causing variants with ultra-rapid nanopore genome

Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol (2018).

Key Publications:
ai.googleblog.com/search/label/health
Google Health Al Blog

github.com/google/deepvariant
DeepVariant on GitHub

DeepVariant and its ecosystem are open-source and actively developed. Find code, documentation, pre-trained models, and key publications at the following resources.
Explore the Technology